- Download PDF
This essay is part of The Launch Sequence, a collection of concrete, ambitious ideas to accelerate AI for science and security. These projects need people to build them. Get in touch.
Summary
America is unprepared to detect new biological threats. Existing pathogen detection methods only identify known pathogens, leaving us blind to novel outbreaks. With frontier AI potentially putting virus design within the reach of more actors, biosecurity is only becoming more urgent.
Within 2–3 years, we could transform our pathogen detection capabilities by adopting new technologies: metagenomic sequencing, which detects both known and unknown pathogens, paired with frontier AI models capable of rapidly analyzing billions of sequencing reads a day. The cost of metagenomic sequencing has been dropping rapidly, making it possible to now collect data at the scale needed to enable AI-powered pathogen early warning. Upgrading US biosurveillance in this way would both provide detailed insights into seasonal pathogen spread in the US and ensure far earlier detection of new outbreaks.
The US should invest up to $100 million per year into constructing a federal metagenomic surveillance system over the next 2–3 years, as the centerpiece of the CDC’s recently announced Biothreat Radar Detection System. Just as the National Weather Service blanketed the country with radars and made its raw meteorological data public, the federal government can generate and share large amounts of metagenomic sequencing data with a turnaround time of 1–2 days, enabling early outbreak detection that could prevent hundreds of billions of dollars in economic damage.
Motivation
Future biological threats
Infectious disease has burdened the world for centuries. The 1918 influenza alone killed up to 100 million people globally. Since then, we have invested billions into infectious disease mitigation, with great progress in developing vaccines, antivirals, and monoclonal antibodies. But over this time, early detection has been sorely neglected, with monitoring of new pathogens like H5N1 still delayed by months.
Our lack of reliable pathogen monitoring is becoming untenable. Rapid progress in AI has many benefits, but it might create a new risk: widespread access to engineered pathogens. Frontier AI systems are becoming rapidly more capable in virology. In a recently published report, 57 expert virologists created 322 multiple-choice questions that test highly advanced virology skills. Recently released AI model evaluations show that frontier systems outperform the vast majority of experts.
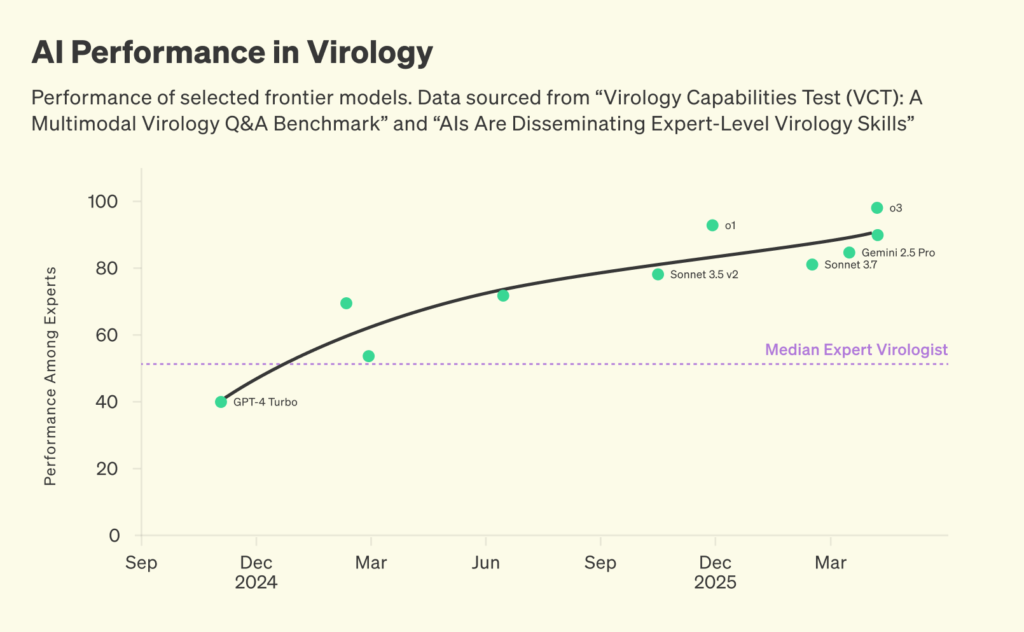
Performance of selected frontier models. Data sourced from “Virology Capabilities Test (VCT): A Multimodal Virology Q&A Benchmark” and “AIs Are Disseminating Expert-Level Virology Skills”
Even AI labs have called for biosecurity measures that go beyond merely securing AI systems themselves. But as AI systems become more powerful, we can also use them to our benefit. Within pandemic preparedness, the most obvious use case is to deploy frontier AI systems for pathogen early detection.
Using AI for biosurveillance
As AI becomes more powerful across domains, many use cases for pathogen early detection will open up. As biological sequence models such as AlphaFold 3 improve, they can predict if unknown DNA or RNA encodes familiar threats. New anomaly detection tools can be trained on incoming data to learn which signals are benign and which are unexpected and potentially dangerous. Finally, AI agents are increasingly able to work across longer time horizons, which should enable them to manually analyze suspicious reads using existing bioinformatic tools.
What would AI-enabled pathogen early detection look like in practice? Imagine billions of sequencing reads coming in daily from airplane wastewater surveillance. An anomaly-detection model, trained on months of historical sequencing data, rapidly scans through these reads and flags a few hundred unusual sequences for deeper inspection. These sequences are then automatically analyzed using a protein-folding model, which predicts the structure of the protein the read encodes. The resulting structural predictions, along with the original sequencing data and associated metadata, are passed to an AI agent that runs further bioinformatic analyses.
While this agent quickly flags most of these sequences as benign, one sequence stands out. Even though its genetic profile doesn’t directly match known pathogens, its predicted protein closely resembles hemagglutinin, the protein influenza viruses use to infect host cells. Recognizing this threat, the AI agent flags the suspicious read for urgent human review.
It’s difficult to predict which AI use case will ultimately prove most useful. But to enable innovation in pathogen early detection in the first place, we need publicly accessible data that companies, researchers, and governments can experiment with. We can learn from weather surveillance on how innovation can be spurred: the government invested in radar infrastructure, made the data openly accessible, and private companies and researchers used it to develop forecasting tools more advanced than what the government could have created alone. The government can play a similar role in pathogen early detection by providing the biological equivalent of weather radar data: metagenomic sequencing data.
The potential of metagenomics
In traditional disease surveillance, we only look for known pathogens. For instance, if a patient arrives with a respiratory infection, we might take a sample from that patient and analyze it using quantitative polymerase chain reaction (qPCR). qPCR uses little probes that can only bind to the genome of a pre-specified pathogen, which is useful for diagnosing known pathogens but frequently misses novel pathogens.
Metagenomic sequencing works differently. Unlike targeted approaches, metagenomic sequencing breaks up all genetic material (DNA and RNA) in a sample into short fragments. Reading out these fragments as sequencing reads, we can identify known pathogens, and further study unknown reads that could represent a new threat. Until recently, the routine use of metagenomics wasn’t feasible due to its cost. However, sequencing is getting orders of magnitude cheaper, and we are now entering a time where sequencing is ready to be deployed at scale.
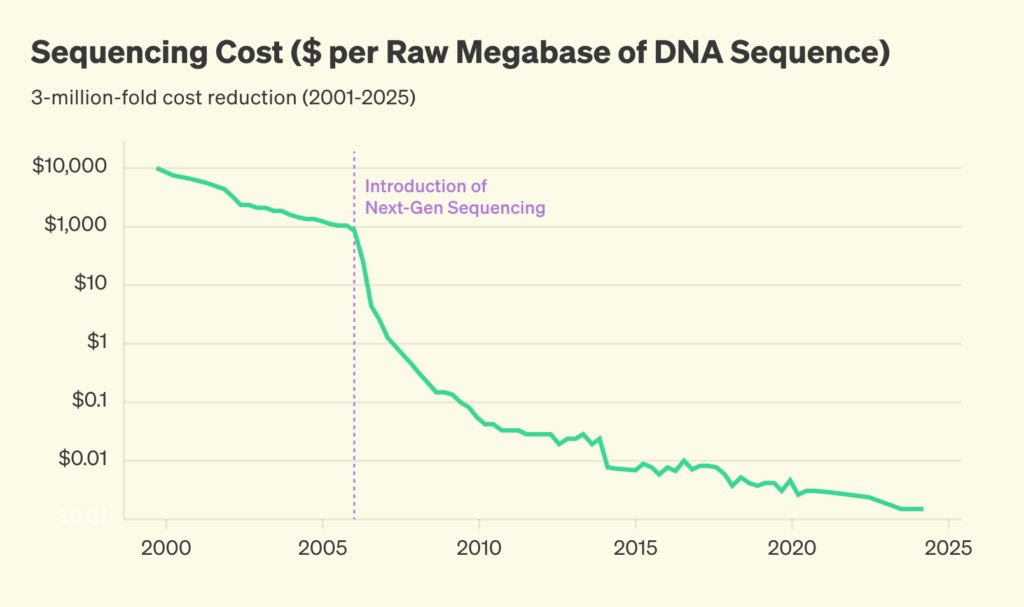
Sequencing cost, inflation-adjusted. Pre-2022 data from Our World in Data; post-2022 data from the Nucleic Acid Observatory. Post-2022 data is based on published sequencing cost data by Illumina. Inflation adjustment is based on quarterly FRED price deflator data.
Bottlenecks for metagenomic biosurveillance
Deploying metagenomic sequencing at scale requires access to many biological samples and sustained funding for sequencing. The US government runs programs that generate several highly valuable sample streams: CDC’s Traveler-based Genomic Surveillance system (TGS) collects nasal swabs and wastewater from thousands of international travelers each week, the Advanced Molecular Detection program (AMD) works with commercial laboratories to analyze clinical samples that test negative for known pathogens, and the National Wastewater Surveillance System collects wastewater across the US covering more than 100 million citizens.
Despite the US government collecting thousands of valuable samples each week, current CDC funding is restricted to targeted assays, leaving metagenomic sequencing largely unfunded. Given the limited commercial incentives to generate and share large-scale sequencing datasets, public investment is required to fill this gap.
Building a metagenomic surveillance system
The CDC should launch an ambitious metagenomic surveillance program, with the explicit goal of generating large amounts of publicly available metagenomic sequencing data. HHS and CDC should ensure that this data is shared with the public. To improve its ability to process this data within the agency, the CDC should partner with AI labs to deploy frontier systems internally, similar to FDA’s recent drive to increase AI adoption. Once metagenomic data streams come online, the Defense Innovation Unit (DIU) should fund organizations to develop advanced analytics capable of rapidly scanning billions of sequencing reads for potential threats.
The required investments in ongoing metagenomic monitoring are modest compared to the large costs of future disease outbreaks. For roughly $100 million per year, about the cost of one F-35 fighter jet, the US could transform its ability to detect new biological threats early. By investing in metagenomic sequencing infrastructure, accelerating the development and diffusion of AI-driven pathogen detection, and ensuring rapid, transparent data sharing, America can significantly strengthen its defenses against future health threats.
Solution
Scaling data generation (2–3 years)
Centers for Disease Control and Prevention (CDC)
As announced in the 2026 President’s Budget, the CDC might receive funding for the launch of a Biothreat Radar Detection System. This new system’s centerpiece should be an ambitious metagenomic sequencing effort.
Within the Emerging and Zoonotic Infectious Diseases division, the CDC should allocate $80 million to build out large-scale metagenomic surveillance capacity.
CDC has several surveillance systems that are particularly well-suited for this purpose. The CDC’s Advanced Molecular Detection System works with private lab companies such as Labcorp to screen laboratory specimens that tested negative for known pathogens, and the TGS program collects nasal swabs and airplane waste from thousands of international travelers each month. Across these systems, establishing metagenomic surveillance would require the following investments:
- Traveler surveillance ($44 million): Expand the TGS program1 to enable nasal swab sampling at 20 US airports. Perform metagenomic sequencing on those samples, using protocols that allow turnaround times of <1 day. This program would allow detection of a new respiratory pathogen before it infected 0.015% of the air traveler population.
- Airplane wastewater surveillance ($10 million): Building on the existing TGS airplane wastewater surveillance program, expand pooled airplane wastewater surveillance to six airports. Covering a different set of pathogens than the traveler surveillance system, detection would be possible before 0.04% of travelers were infected.
- Respiratory sample surveillance ($26 million): Expand on AMD’s existing partnerships with large commercial laboratories by running pooled metagenomic sequencing on PCR-negative respiratory samples. This would enable rapid detection of pathogens with fast disease onset.
In setting up its metagenomic surveillance system, the CDC should use technologies that allow fast turnaround times. This includes using sequencing technology that allows sample-to-data turnaround times of under 24h for swab samples, and under 48h for wastewater (Appendix).
To maximize the benefit of large-scale metagenomic data generation, actors beyond the CDC, such as innovative companies and nonprofits, need data access. To this end, the CDC should allocate funding for purchasing data storage that allows for data to be stored and shared quickly (publication within 24 hours of data generation), working both with the Sequencing Read Archive and commercial cloud providers.
Department of Health and Human Services (HHS)
Standards need to be established to allow the generated metagenomic sequencing data to be promptly shared. To facilitate this, HHS should provide guidance on data sharing. This guidance would establish how human genomic material should be removed prior to upload and would mandate the provision of certain types of metadata (time, location).
Scaling data analysis (2-4 years)
As metagenomic data streams start coming online, we need adequate analytics to parse incoming data. This will involve an ecosystem of companies and nonprofits that can leverage frontier AI models to detect new pathogens. To support the development of this ecosystem, agencies should take the following steps:
Defense Innovation Unit (DIU)
Large amounts of metagenomic sequencing data can be used to develop sophisticated threat detection algorithms, such as those identifying genetic engineering signatures or novel pathogen anomalies. But current methods to screen sequencing data for threats were often designed for smaller datasets (for instance, analyzing individual respiratory samples in forward-deployed settings). To support the routine analysis of billions of sequencing reads a day, we need a new set of detection methods. Building on prior work to advance metagenomic analysis of wastewater, DIU should fund a further set of organizations to develop AI-based threat detection methods that enable analysis of large amounts of metagenomic sequencing data.
Further resources
- CDC, SARS-CoV-2 Sample Positivity in Travelers Can Predict Community Prevalence Rates: Data from the Traveler-Based Genomic Surveillance Programme, 2024.
- Defense Innovation Unit, DIU Demonstrates Capability To Find Novel Threats in High Complexity Wastewater Data, 2025.
- Nava Whiteford et al., Towards Ubiquitous Metagenomic Sequencing: A Technology Roadmap, n.d.
- Nucleic Acid Observatory, n.d.
- CDC, Advanced Molecular Detection (AMD), n.d.
- CDC, Traveler-based Genomic Surveillance (TGS), 2025.
- HHS, FY 2026 Budget in Brief, 2025.
- White House, Technical Supplement to the 2026 Budget, 2025.
Appendix
Technical details of metagenomics
Unlike targeted approaches such as antigen tests or quantitative polymerase chain reaction (qPCR), metagenomic sequencing works by breaking up all genetic material (DNA and RNA) in a sample into short fragments, reading the sequence of DNA/RNA bases in each fragment, and then matching these reads against reference databases to determine which organisms they came from. The cost of sequencing has dropped by several orders of magnitude in recent years.
Depending on the sample type, sequencing methods should differ correspondingly. For high-throughput swab surveillance, Oxford Nanopore Technologies (ONT) sequencing is ideal because sample preparation and sequencing can be accomplished in under 24h. In contrast, wastewater samples contain lower pathogen concentrations, requiring higher read depth. For this use case, short-read Illumina platforms (e.g., NovaSeq X 10B) deliver the necessary output within a 48-hour window.
-
Numbers are based on HHS estimates and system cost & detection sensitivity simulations. Please contact the author for more details.